Res Area 1: Molecular genetic manipulations
Brief description: Over teh years we have developed a variety of Cre recombinase - and Dre - recombinase drivers and reporter lines, based on the concept that single cell type manipulations will not be posiible by using individual genetic elements, but rather intersection of two or more gene expression domains. In addition, we created unique conditional knock-in reporter strategies that result in teh removal of an endogenous target gene and its replacement with a histochemical reporter (Alkaline Phosphatae - AP) or eGFP (enhanced Green Fluorescent protein). By using this strategy, we can reveal the cell autonomous effect of gene manipulation in addition to characterization of the cell type distribution of the targeted genes.
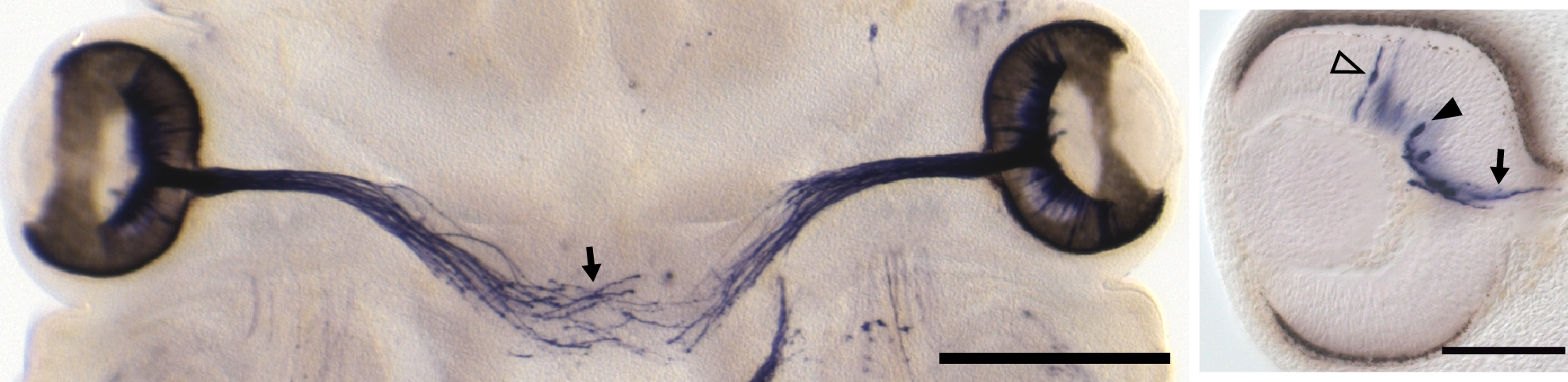
Embryonic day 12 RGCs leaving the eye (right) and crossing the midline at the optic chiasm (left)
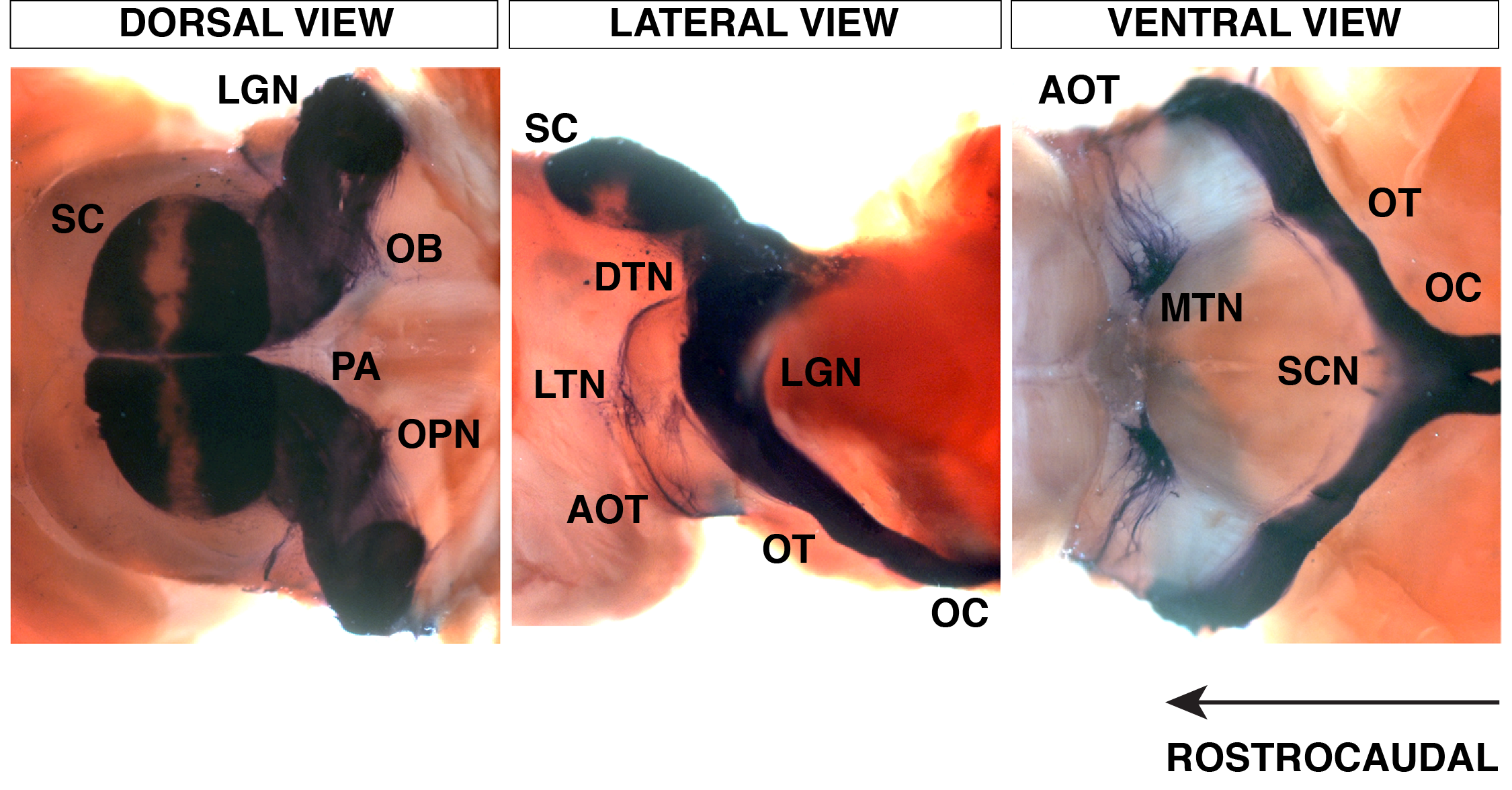
RGC axons innervating retinorecipient areas of the brain
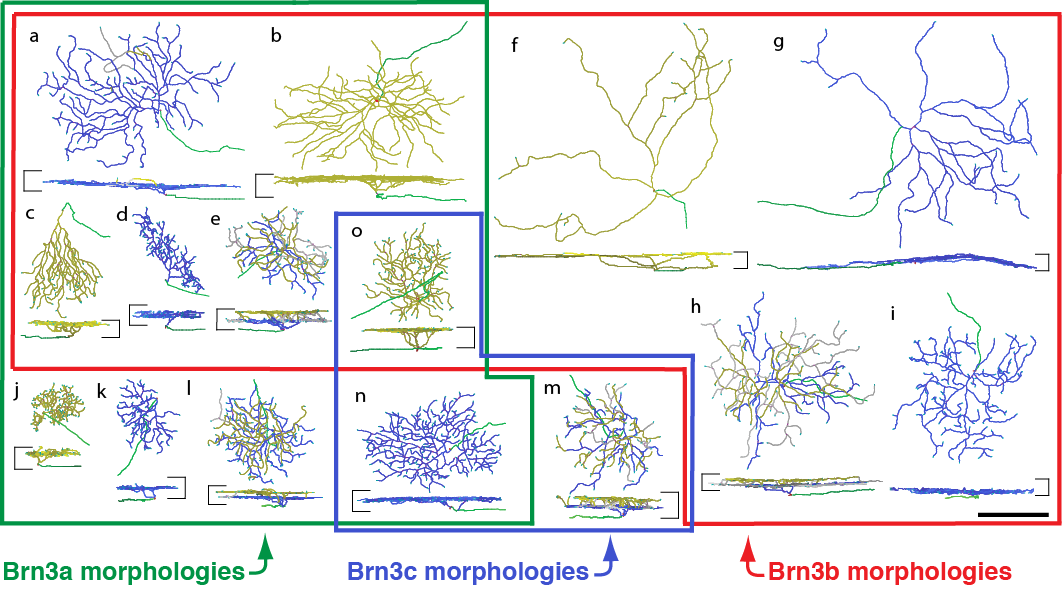
Dendrite Morphologies of RGCs expressing Brn3 transcription factors
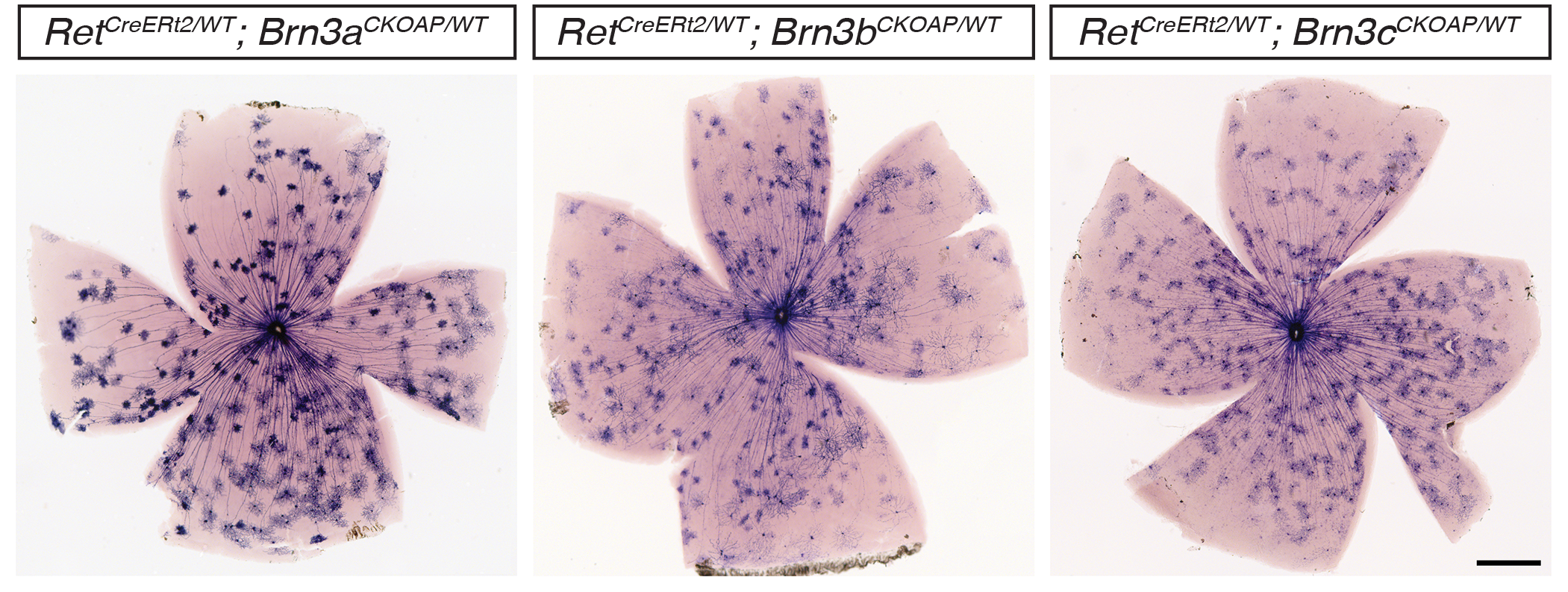
RGC types expressing cRet in combination with Brn3 transcription factors
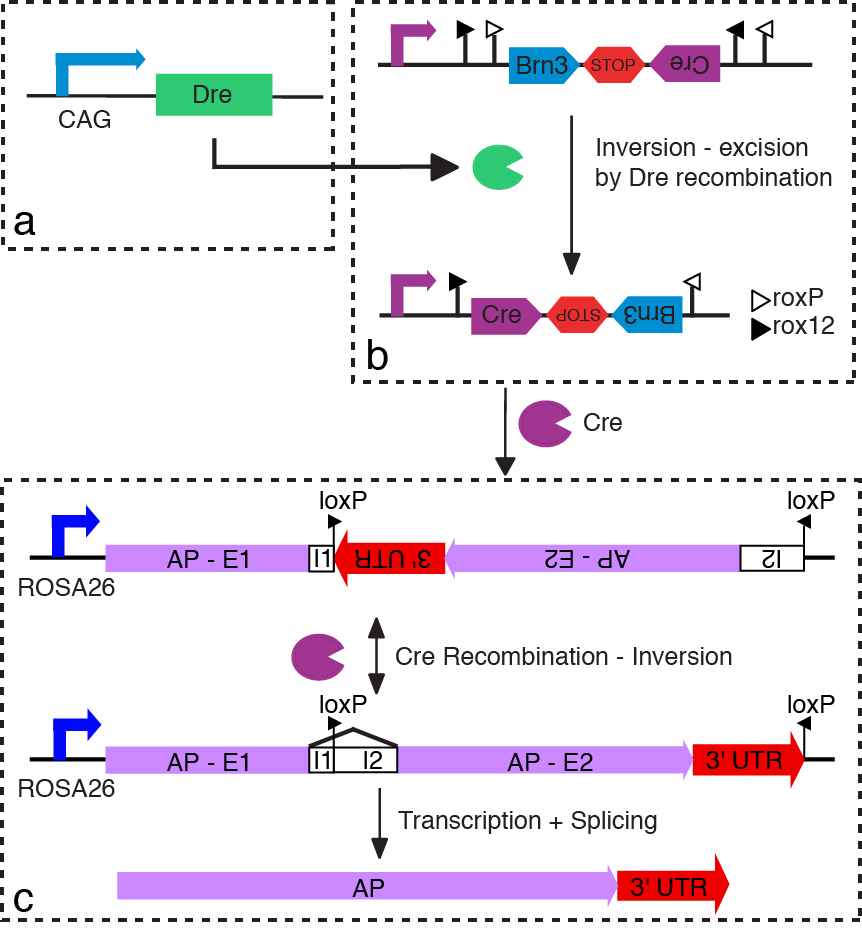
Intersecting three different genetic loci using site specific recombination. The first locus drives Dre recombinase, a second locus drives Cre recombinase in a Dre dependent manner, and a third locus reports Cre activity, in the cell expressing all three loci.
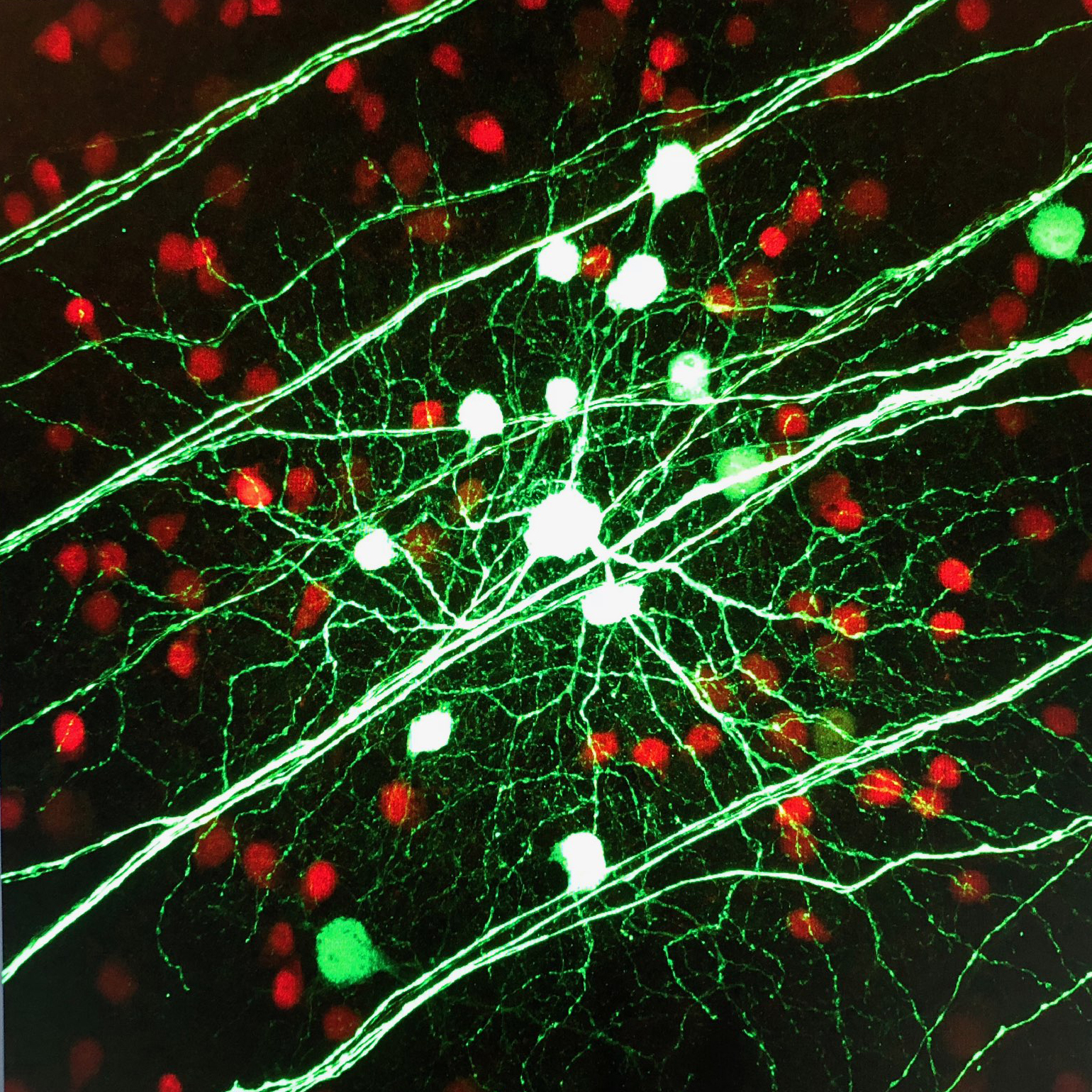
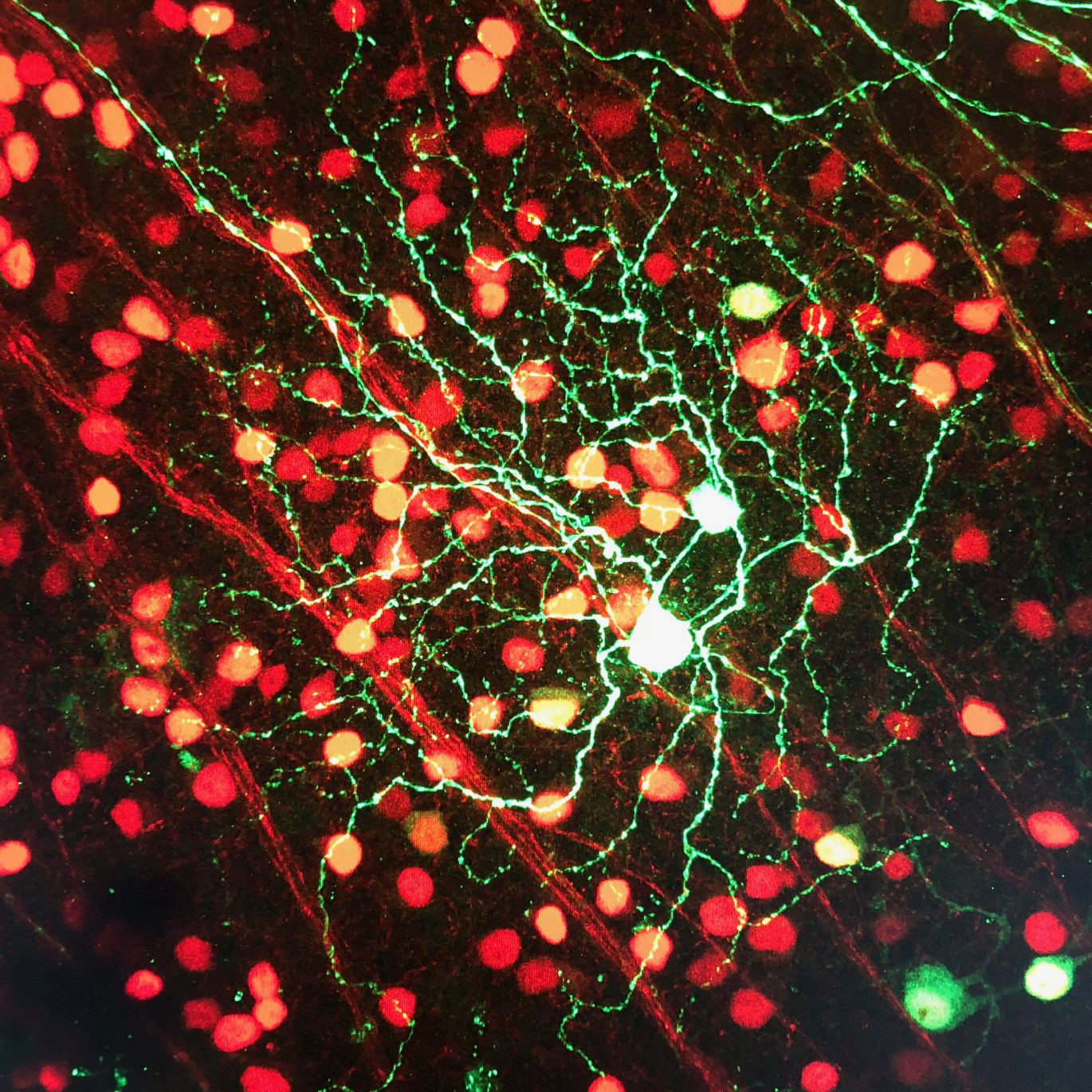
Brn3c-Cre expressing ON alpha and ON-OFF DS RGC infected with FLEX-eGFP AAV1
Res Area 2: Neuronal Transcriptomes
We use immunomagnetic or fluorescence activated cell sorting in order to isolate and purify neurons expressing our genes of interest. We can therefore study the transcriptomic effects of gene ablation with single cell resolution. In 2017 we identified an extensive set of genes expressed in RGCs and regulated by Brn3 transcription factors. These genes encode further transcription factors, cell surface molecules (adhesion, axon guidance and signaling receptors), and many other interesting targets with functions in neuronal morphology.
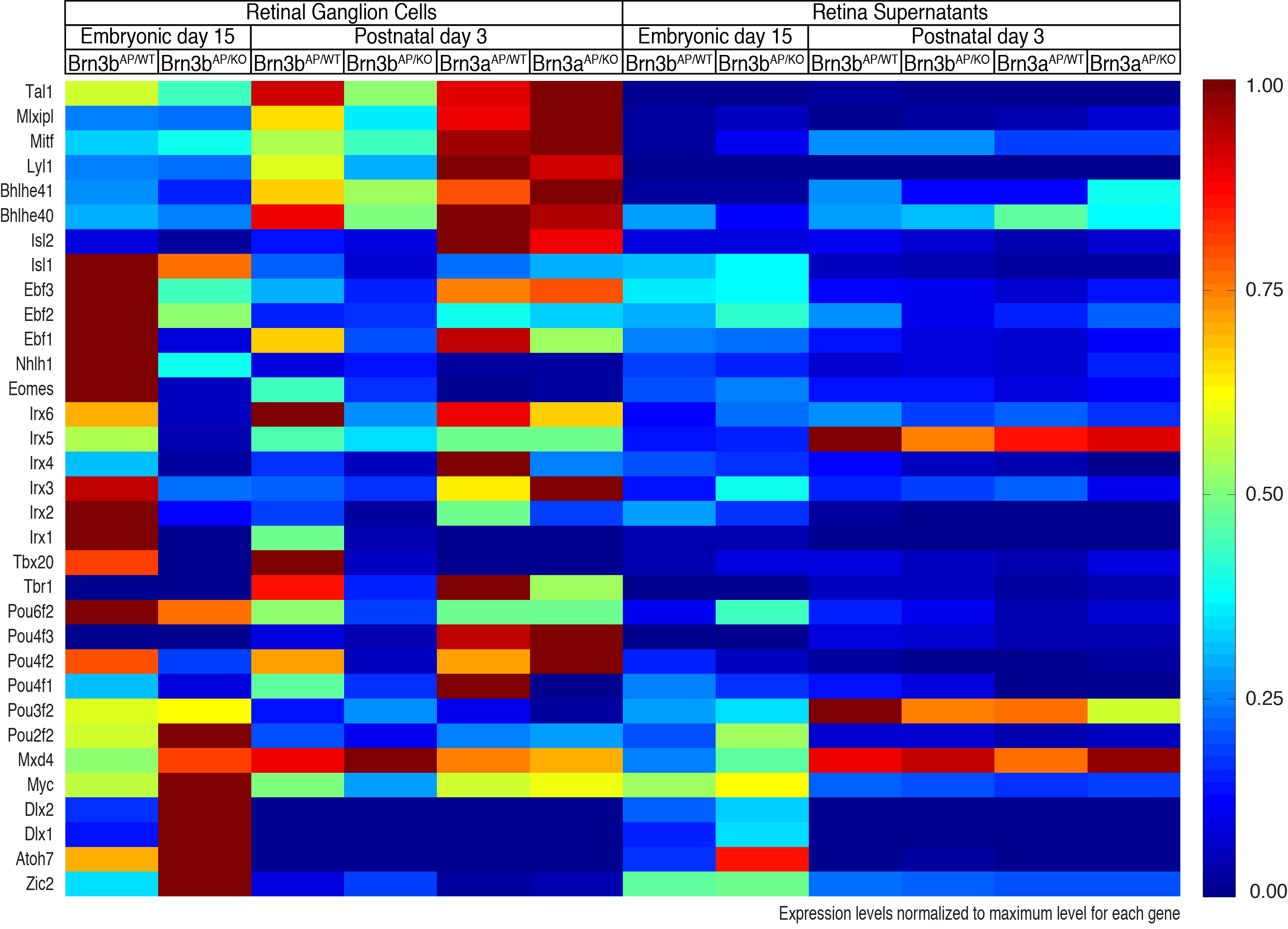
Selected transcription factors expressed in RGCs and retinas and differentially regulated by Brn3 transcription factors.
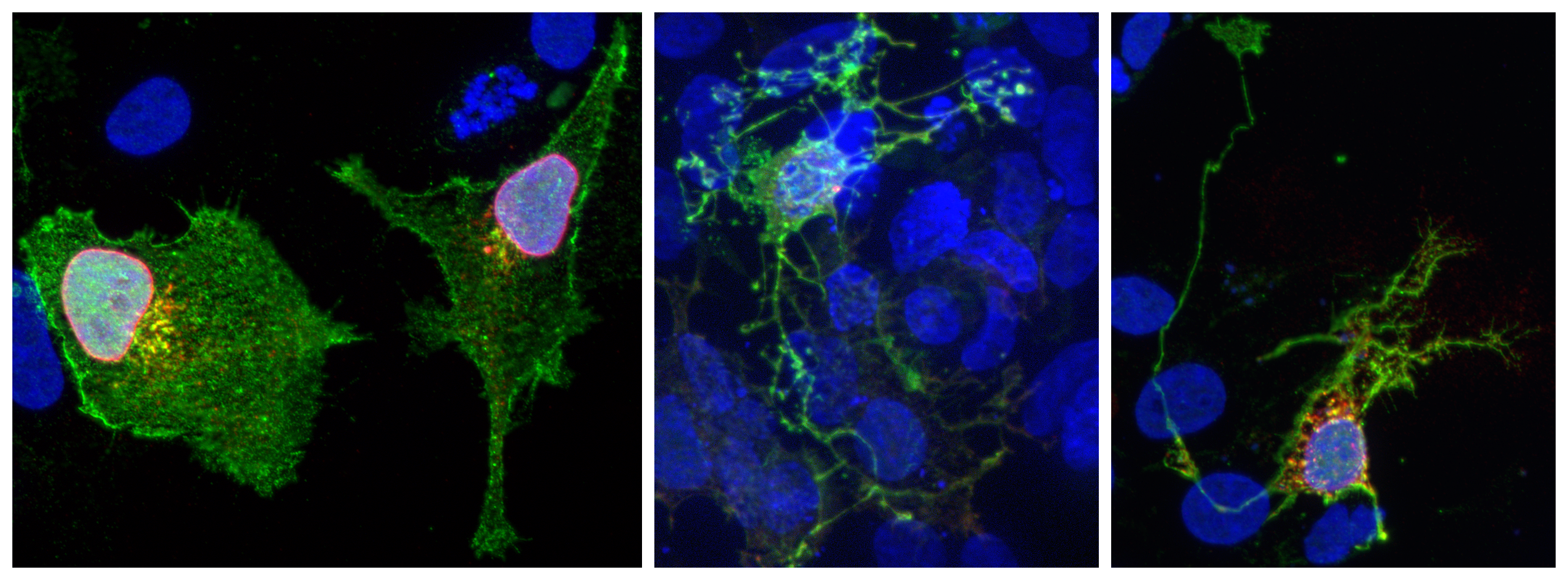
HEK293 cells overexpressing genes Brn3 transcriptional target genes expressed in RGCs and believed to influence neuronal morphology.
Res Area 3: Visual Physiology/Behavior
In order to analyze the effects of genetic manipulations on RGC physiolgy and visual function, we continue to develop software and hardware solutions for retina electrophysiology and visual reflex testing. We present the retina ex vivo with visual stimuli testing the major physiological RGC types, and then use multi-electrode array recordings in order derive input - output functions, under wild type or knock-out or manipulated conditions. In parallel, we use tests for visual reflexes in order to ascertain the consequences of our genetic manipulations on specific visual functions.
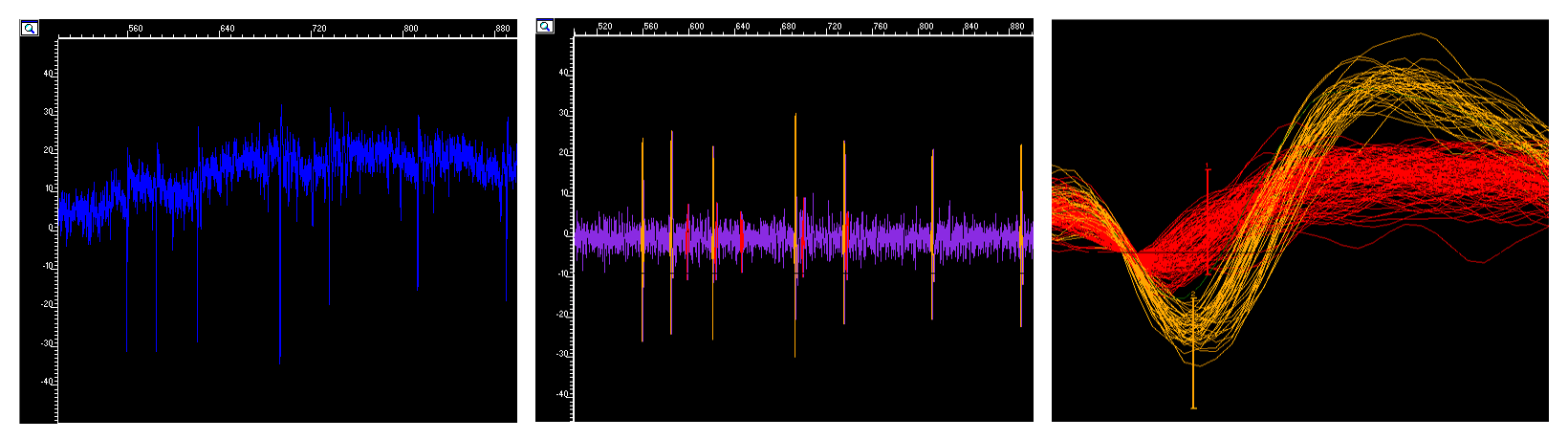
Example of a multielectrode channel detecting spike trains from two neighboring RGCs. Top to bottom: (i) Raw trace, (ii) Filtered trace with detected and sorted Spikes from the two RGCs, (iii) Sorted, and clustered overlayed spike cutouts color coded by RGC cluster.
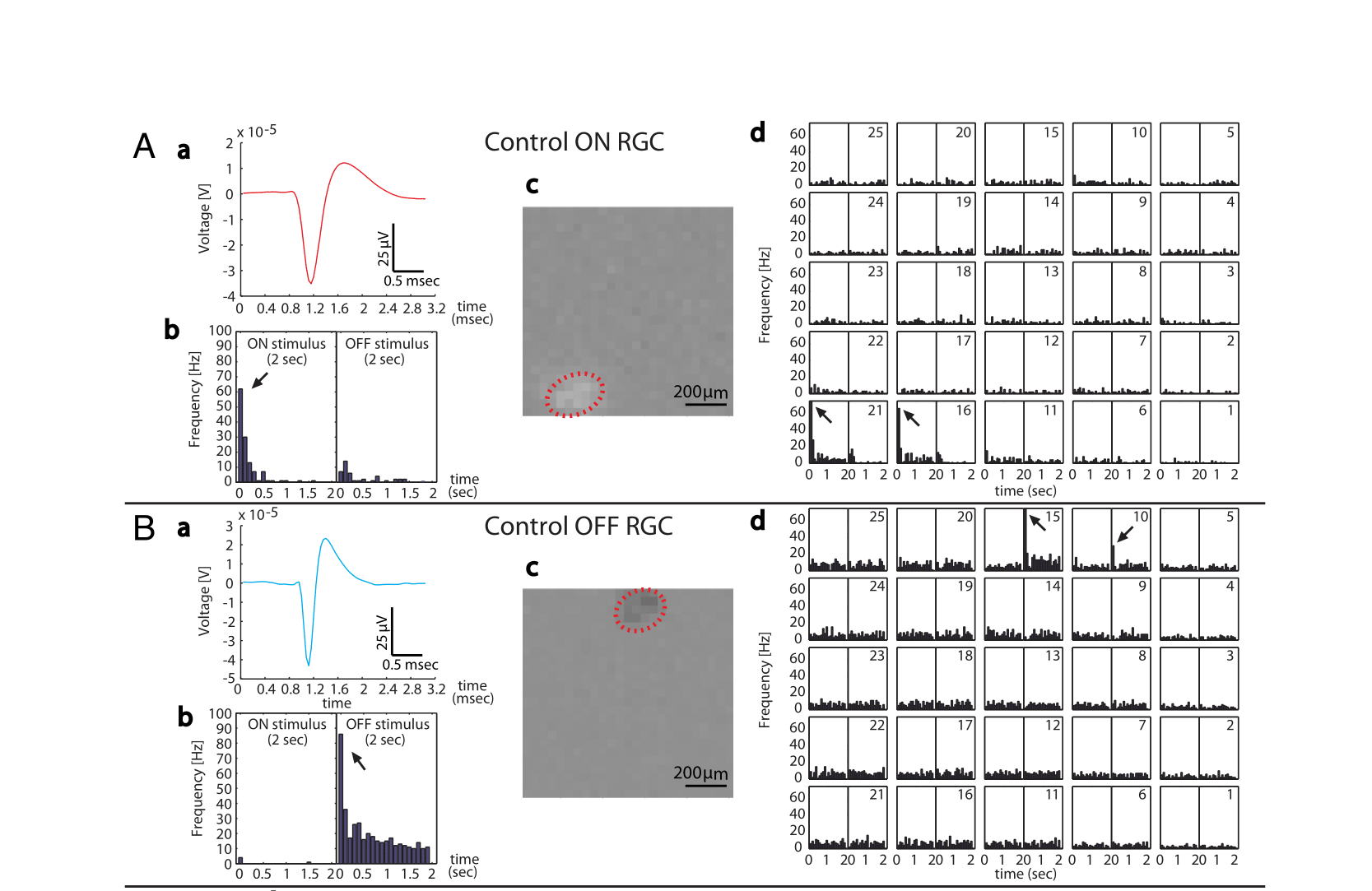
Examples of receptive field properties of “ON” (top) and “OFF” (bottom) RGCs, as revealed by multielectrode array recordings of retinas exposed to full field (b), checkerboard (c) or tiled spots (d) stimuli. The action potential template for each cell is shown in a.

Example of CMOS MEA recordings from a WT retina. This array can capture 4000 channels simultaneously.
WT mouse fleeing to a refuge in response to a looming stimulus, mimicking an approaching aerial predator.
WT mouse freezing in response to a sweeping stimulus, mimicking an aerial predator flying overhead.
Online live detection of a mouse tracking a sinusoidal grating moving horizontally. Optomotor responses can be observed.